COMPUTATIONAL BIOLOGY: |
||
Principal Investigators: Larry Forney (UI), Steve Krone (UI), Paul Joyce (UI), Frank Gao (UI)
Numerous mathematical models have been developed for the biocomplexity project to provide insight regarding the adaptive evolution and speciation of prokaryotes, the biogeography of bacterial populations, and the spatial and temporal dynamics of microbial communities— particularly those within biofilms. This page contains some of the highlights of the computational biology research and provides additional links to more information on Biofilm Models and Microbial Community Spatial Simulators. Spatial Variation in Microbial Community Structure (...more) To assess "between location" variation in microbial community composition, matrices of tiles (5 x 5 one-inch tiles) were placed at 4 locations within the stream emanating from Mickey Hot Springs that vary in temperature by ~6 °C. Since the time they were deployed (August 2003), replicate samples (N=5) were removed from each grid on 2 occasions using a randomized sampling scheme. Total community DNA has been isolated from the samples that will be analyzed by determining profiles of 16S rRNA gene terminal restriction fragment length polymorphisms.
The spatial variability "within location" was assessed by removing two entire 5 x 5 tile grids from two distinct locations. The biofilms from each tile were separately removed and analyzed by determining profiles of 16S rRNA gene terminal restriction fragment length polymorphisms (T-RFLP). Each sample was analyzed using two restriction enzymes. Statistical analyses of the data showed that there were no meaningful differences among the communities on the tiles. This result was somewhat surprising since growth on the tiles was obviously patchy. While it is possible that the analyses done failed to distinguish differences between tiles, it seems more likely that the spatial heterogeneity that was visually apparent occurs at a scale less than 1 x 1 inches, and that each 1 x 1 inch space has roughly the same species diversity. The species composition of these communities is being determined by construction and analysis of 16S rRNA clone libraries. *** Read on to learn about Stochastic Spatial Modeling of Microbial Communities. |
Biofilm Studies (...more)
Studies to understand the physiological and genetic variability within microbial biofilms have been initiated using the model organism Escherichia coli. These studies have three aims: (1) characterize the genetic and physiological characteristics of single species biofilms; (2) establish theoretical and/or conceptual models for physiological and genetic responses in biofilms; (3) extend the models to multispecies biofilms such as those found in geothermal springs. Experiments have been done using two basic strategies: (a) monitoring the expression of specific genes in situ using transcriptional fusions to fluorescent reporter genes; and (b) the analysis of genotypic and phenotypic clonal variation in mature biofilms. The former relies on confocal microscopy to visualize fluorescent cells at various depths within intact biofilms. The latter employs histological methods to obtain thin sections (100 µm) from which clones can be isolated, and their characteristics determined. An experiment was performed to assess adaptive evolution within the biofilm. Three-month old biofilms of E. coli were sectioned and clones were isolated. For each clone, the mutation frequency (streptomycin resistance) and specific growth rate were determined. It was expected that the growth rate of clones would not vary but that the mutation frequency would increase with depth. Surprisingly, we found that the both the mutation frequency (Figure 1a) and growth rates (Figure 1b) showed a bimodal distribution. The maximal mutation frequencies were found in the middle depths and at the bottom near the substratum, and there was an inverse relationship with the specific growth rate of clones. 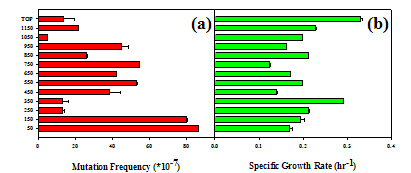 These data indicate there is rapid diversification of clones within the biofilm to local conditions that exist within microenvironments, and that perhaps an increase in mutation rate is accompanied by a decrease in the fitness of clones (as measured by differences in growth rate) due to the accumulation of deleterious mutations. *** For more information on biofilms, click HERE. |
Life at Interfaces: Biocomplexity in Extreme Environments |
||
 |
||
|
||
|
|
||
| last update: June 2006 | webmaster: jhinds@uidaho.edu | ||
