COMPUTATIONAL BIOLOGY (Spatial Simulations): |
||
Modeling Microbial Community Network Interactions
Principal Investigator: Steve Krone (UI) The goal of this research is to understand the mechanisms of microbial ecology and evolution, both generally and in the particular setting of rock-water interfaces associated with hot springs . Our basic approach is to develop mathematical models that characterize essential features of microbial systems. One of the key issues we are addressing is the amount and kind of biodiversity in such microbial systems. It has been demonstrated in recent years that the explicit role of space coupled with stochastic effects on various length and time scales have a profound influence on the generation and maintenance of high levels of diversity. Our models (interacting particle systems) take into account all such aspects of the dynamics. In addition, one of the things that distinguish microbial diversity from more traditional ecological systems is that evolutionary changes can occur on the same time scale as ecological changes. This, when coupled with the high degree of variability in microbial genomes, leads to important sources of diversity. Through our research we developed Stochastic Spatial Simulator (see image to the right), a Windows-based program for simulating ecological and evolutionary systems that take into account explicit spatial and stochastic dynamics. The software is freely available on the web at: http://www.webpages.uidaho.edu/~krone/winsss/sss-main.htmlThis program simulates the evolution of complex microbial community interaction networks, while keeping track of things like the degree of connectivity in the graphical network, the types of interactions that are stable (competition, predator-prey, mutualism), and the effect on species densities. Using our stochastic spatial simulator, we are analyzing a collection of models that involve cyclic successional dynamics. Such models are appropriate for microbial systems that are spatially structured (e.g., within a biofilm) and where degradation of nutrients occurs in a symbiotic manner. These dynamics lead to fluctuations in species and resource densities that promote diversity. We have shown that spatial structure in these systems leads to coexistence and stable oscillations that are not present in a well-mixed liquid environment. The complex spatio-temporal patterns that arise are similar to those studied intensively by mathematicians in recent years and our models provide some insight into what causes such patterns. Our studies are leading to an understanding of the way microbial communities organize themselves and the consequences for diversity and stability. |
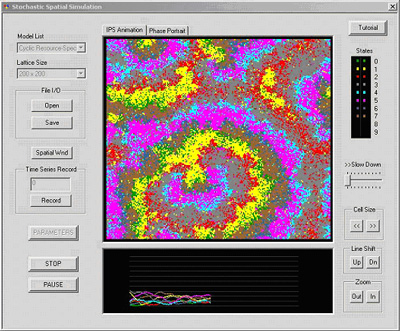 Stochastic Spatial Simulator RELATED PUBLICATIONS Gao, F., Zhong, X., La, H.-J., Krone, S., and Forney, L. (pending). A 3-D multispecies biofilm model using energy allocation. Biotechnol. Bioeng. Guan, Y. and Krone, S. 2004. WinSSS: Stochastic Spatial Simulator. Bulletin of Ecological Society of America 85(3): 102-104. Guan, Y., Fleißner, R., Joyce, P., and Krone, S.M. 2006. Markov Chain Monte Carlo in Small Worlds. Statistics and Computing 16(2): 193-202. doi:10.1007/s11222-006-6966-6. Krone, S.M. 2004. Spatial Models: Stochastic and Deterministic. Mathematical and Computer Modelling 40(3-4): 393-409. Labrum, M., Soule, T., Blue, A., and Krone, S.M. 2004. On the Evolution of Structure in Ecological Networks. Proceedings of 4th International Conference on Computational Science (ICCS), Bubak, M., Albada, G.D.v., Sloot, P.M.A., Dongarra, J.J. (Eds.), Kraków , Poland , June 6-9, 2004 . |
Life at Interfaces: Biocomplexity in Extreme Environments |
||
 |
||
|
||
|
|
||
| last update: June 2006 | webmaster: jhinds@uidaho.edu | ||